Information about LIBNA.
 Laboratory of Integrated Biomedical Micro/Nanotechnology and Applications’ (LIBNA) vision in integrating biology and medicine with micro and nanotechnology can be categorized into two broad areas, namely how micro/nano-fabrication can help solve problems in life sciences (such as diagnostics, therapeutics, and tissue engineering), and how we can learn more from life science to solve important problems in micro/nano-science and engineering (such as bio-inspired self-assembly, etc.).
Laboratory of Integrated Biomedical Micro/Nanotechnology and Applications’ (LIBNA) vision in integrating biology and medicine with micro and nanotechnology can be categorized into two broad areas, namely how micro/nano-fabrication can help solve problems in life sciences (such as diagnostics, therapeutics, and tissue engineering), and how we can learn more from life science to solve important problems in micro/nano-science and engineering (such as bio-inspired self-assembly, etc.). 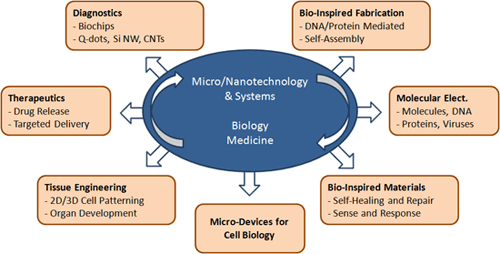 Thrust areas of research in our group are:
Thrust areas of research in our group are:
- Field Effect Transistor Biosensing
- Solid-State Nanopores
- Point-of-Care Microfluidic Diagnostics Systems
- Resonant Mass Sensors for Cellular Analysis
- 3D Bio-Fabrication
This is schematically shown to the right.
Our Principal Investigator is Rashid Bashir.
Check out the tabs below or on our website here to learn more.
Detection of Viable Pathogens Using Label-Free Electrical Detection of Nucleic Acid Amplification
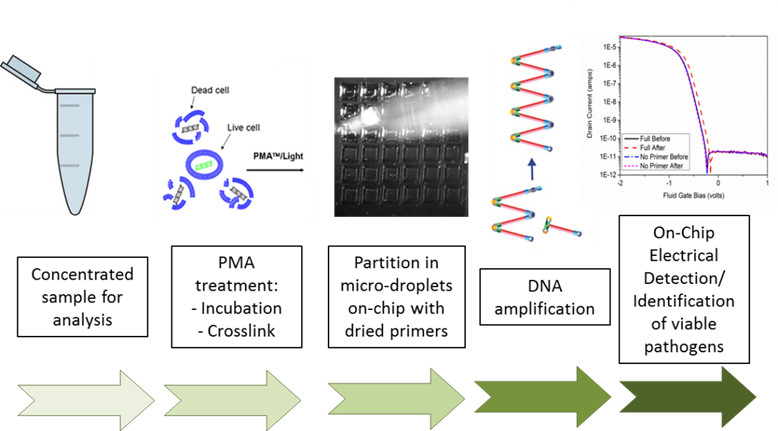
Leveraging developments on electrical label-free detection of amplification and expertise in microfluidic systems, we plan to continue the development of an automated microchip for the detection of pathogens. Our group has previously demonstrated specific detection of foodborne pathogens in a silicon microarray. A primer dehydration protocol enables the detection of multiple genes when the sample is partitioned in micro-reactors with different primer sets. In addition, we have shown that the amplification reaction can be electronically detected using ion-sensitive field-effect transistors that monitor the electrolyte acidity. We now look to take advantage of the intrinsic scalability of electronics and the arraying technologies that we have developed to do electrical detection of multiple reactions.
This will require creating platforms for parallel measurement of transistors and the development of a robust on-chip reference electrode that overcomes the current requirement of having an off-chip electrode which is limiting the number of parallel reactions that can be performed. Additionally, the incorporation of a new pre-amplification step will discriminate live vs. dead pathogens. We will study the on-chip performance of viable qLAMP and the viability of performing the treatment on a microfluidic device. In this way, using electronics and microfluidics, we will develop a DNA amplification system for the detection of foodborne pathogens outside a laboratory setup.
Improving the sensitivity of field-effect biosensors through electronic desalting
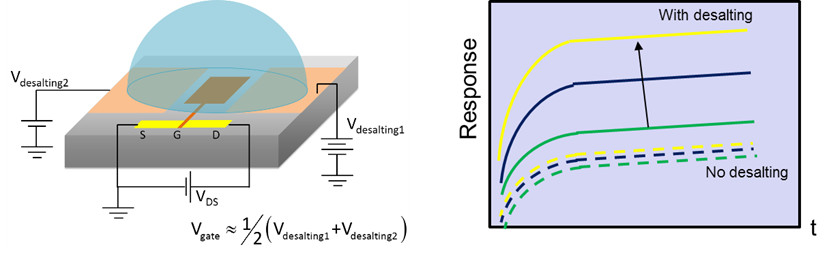
Direct molecular sensing in nano-bio sensors facilitates a label-free electrical detection route, which greatly simplifies the diagnostic instrumentation, useful for point-of-care applications and resource-limited settings. Nanowire-based FET devices have shown potentially very high sensitivity for the detection of small biomolecules, such as nucleic acids, proteins, and viruses. FETs being highly scalable, they are ideally suited for miniaturizing the diagnostic platform.
However, the inability to commercialize these sensors is largely because we cannot sense directly from blood or serum due to shielding of molecular charge by the excess of ions. Screening by background salt ions interferes with sensing from physiological media by decreasing the apparent charge at the sensor and reducing the overall sensitivity. Our goal and design is an electronic desalting scheme, using on-chip polarizable electrodes around a FET that locally deplete the salt ions to overcome shielding, that improves the sensitivity to detect bound molecules (e.g., miRNA cancer markers). By overcoming ionic shielding, charge transduction to the FET and, consequently, the sensitivity can be maximized. The on-chip electrodes also gate-bias the FET for simultaneous sensing during desalting, to beat the competing back-diffusion of the salt ions. The entire sensor is miniaturized to the sub-nanoliter scale of a FET-in-a-droplet, which can pave the way for multiplexed electronic detection with high sensitivity from physiological media.
Biosensing on Large-Scale Field-effect Transistor Array
Diagnostic techniques have become critical to modern health care not only for patient diagnosis and optimization of treatment, but also for providing vital information regarding pathways for complex diseases. Ideal biosensors provide the maximum desirable data with the least amount of complexity. However, to date, most biosensors either yield very specialized data or are very complex and expensive. Researchers have attempted to address this need with the development of point-of-care sensors that can provide accurate data without the need for expensive equipment. Semiconductor manufacturing techniques offer a particularly attractive opportunity to design a biosensor that is highly versatile; extremely inexpensive; produced with high yield, stability, and reproducibility; and highly sensitive to target analytes due to device size. Such fabrication techniques at industrial class foundries have been honed to perfection with optimization and standardization. Prototyping of new device designs in a foundry can critically enable the rapid commercialization of new bio-medical platforms.
We are working with a versatile electrical biosensor platform consisting of tens of thousands of devices fabricated with a silicon-on-insulator line. The platform consists of a unit cell transistor integrated seamlessly with control and read-out circuitry that is amenable for immediate commercialization. Each cell consists of a sub-micron FET Sensor with a near-Nernst pH sensitivity of around 56-59 mV/pH and a resolution of <0.01 pH. The gate oxide is directly exposed to the target analytes, instead of to the commonly employed floating gate architecture. This enables many critical advantages, including increased sensitivity due to the elimination of parasitic coupling capacitances and reduced vulnerability to crippling factors such as electrostatic discharge. The unit cell can be coupled to a variety of different surface chemistries for different target analytes and applications.
Stacked Graphene Nanopores to control DNA transport
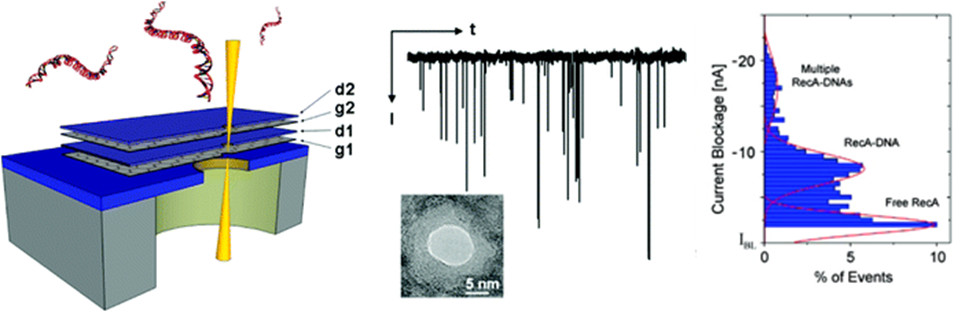
Nanopore based DNA sensing methods use electrophoresis to drive negatively charged DNA molecules through nanometer-sized pores and monitor the change in ionic current to examine the length and sequence of DNA molecules. It is an inexpensive and attractive alternative to traditional sequencing and analysis technologies as it is a label-free, amplification-free, single-molecule approach that can be scaled for high-throughput DNA analysis. Our research involves the application of TEM-drilled nanopores in membranes of sub-nanometer thick graphene combined with robust dielectric materials. We have demonstrated the sensing and differentiation of DNA and RecA-DNA complex transport through stacked layers of graphene and Aluminum oxide. Current goals include exploiting these unique structures to control/reduce DNA transport rate as well as nanopatterning of graphene to enable DNA sensing through graphene conductivity fluctuations. The aim is to improve the reliability and sensitivity of nanopore-based DNA sensing
Detection and Quantification of DNA Methylation
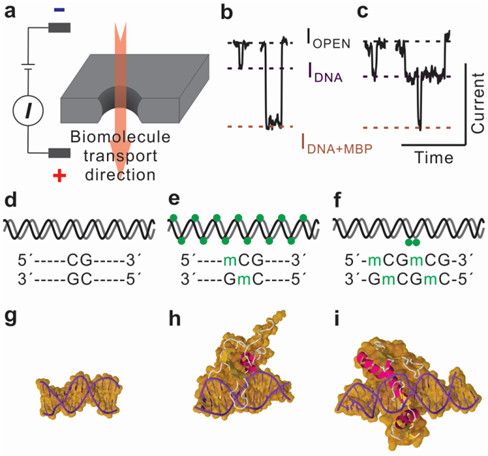
DNA methylation, an epigenetic modification of a methyl group at the 5-carbon position of cytosine (5mC), has been found to be closely associated with carcinogenesis. Thus, genomic extracts holding this modification can be used as biomarker for sensitive detection of cancer, and these genomic DNAs can be collected from serum, plasma, urine, and stool. Nanopore-based direct methylation assay circumvents bisulfite conversion and PCR, suitable for screening of low sample volume obtained from genomic extracts. Methylated DNA is selectively labeled with methyl-binding proteins. Nanopore assay demonstrates discrimination of hypermethylated and unmethylated DNA on various length DNA fragments using sub 10 nm nanopores. Furthermore, low-methylated DNA fragments are differentiated from unmethylated DNA and the position of methylation is profiled using sensitive nanopore-based methylation assay.
Concentration and analysis of circulating DNA
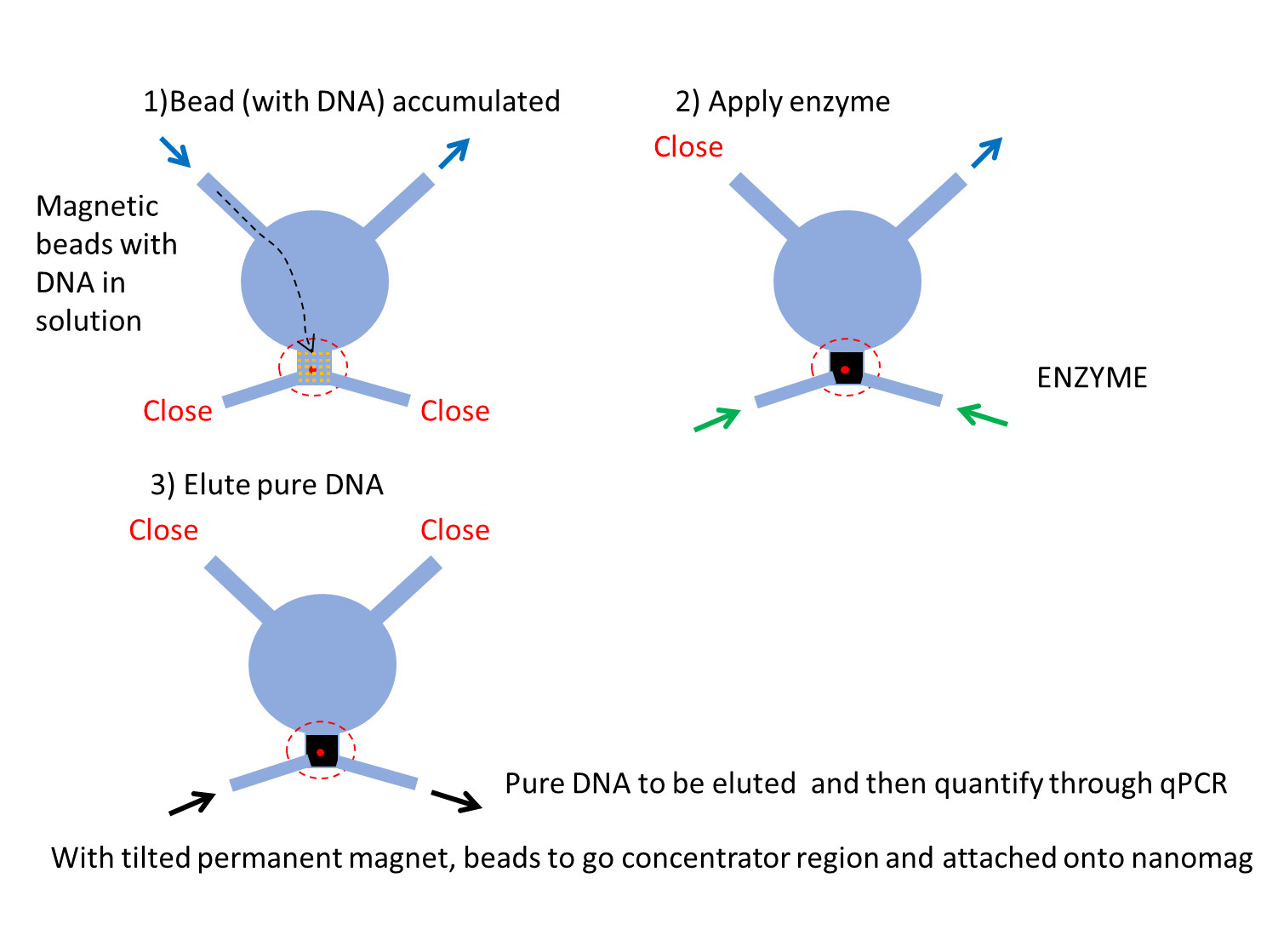
We are developing protocols and apparatus for concentration and analysis of circulating DNA derived from cancer cells using magnetic beads and microfluidics. Circulating free DNA in cancer patients may be a potential indicator to monitor cancer progression and characteristics. For example, CpG sites in Circulating free DNA may be good prognostic or predictive biomarkers, sensing nanopore devices to measure nano electric signals. To do that, DNA molecules of short fragments, which may exist in a low frequency of clinical specimens, should be highly concentrated so that DNA can be detected with more reliable sensitivity and at fast speed. Currently, I have been working on making a great DNA concentrator with nanomagnets, patterned with nickel, soft magnet materials. Thin nanomagnets can exhibit enhanced magnetic flux, which can be generated by a permanent magnet. Combining this nanomagnet with microfluidics may provide a great benefit to capture with an extreme rate of concentration of magnetic beads, which can be bound with short oligo DNAs. The chemistry of DNA binding to the beads and removal from beads is another job for me to work for DNA concentration method.
Quantification of Viral Load Using Liposome Encapsulation for HIV Detection
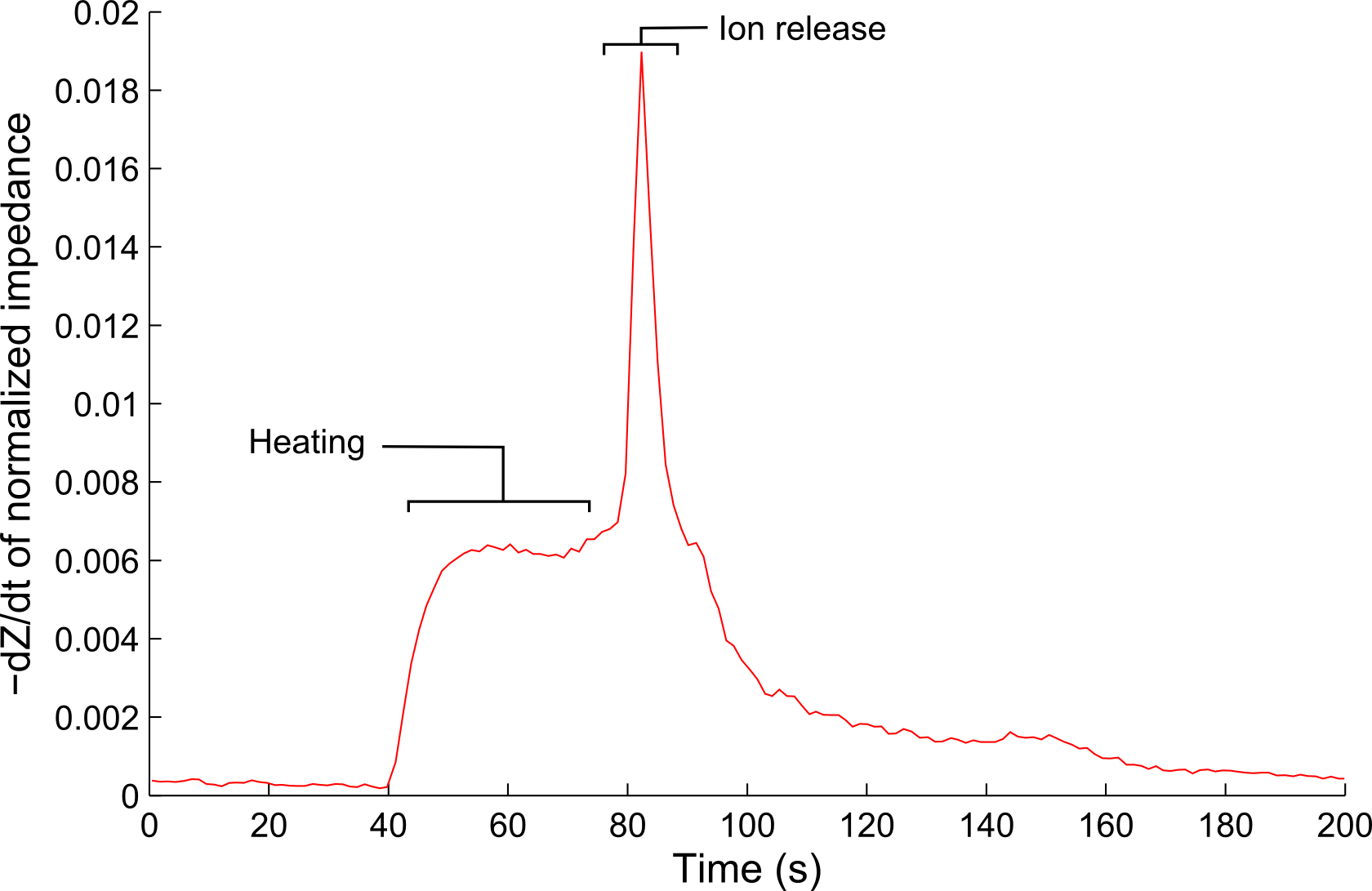
We are developing a technology based on Ion-Release Impedance Spectroscopy (IRIS), a technique that employs liposomes as a biosensor component for the detection of small biological entities. This technique relies only on electrical impedance measurements, eliminating the need for incorporating cameras, lasers, and other optical components which often increase the size and cost of a biosensing platform.
The overall approach of the technique is as follows:
- The analyte of interest is separated from a biological sample by immunoaffinity chromatography or some other purification/isolation method in a microfluidic device.
- The captured analyte is tagged with a liposome encapsulating ions (such as 10X PBS) which are functionalized on its surface with an antibody or peptide that will specifically bind the analyte.
- Unbound liposomes and background media are replaced with low-conductivity media, such as deionized water.
- The microfluidic chamber is heated, which increases the fluidity of the liposome membrane, eventually causing it to release its contents.
- The change in electrical impedance is measured throughout the process, and a decrease in impedance is observed as a result of the addition of ions into the media.
- The number of liposomes in the microfluidic chamber can be determined from the impedance change that is measured, and is the basis for quantifying the analyte.
Blood Cell Counting at Point-of-Care
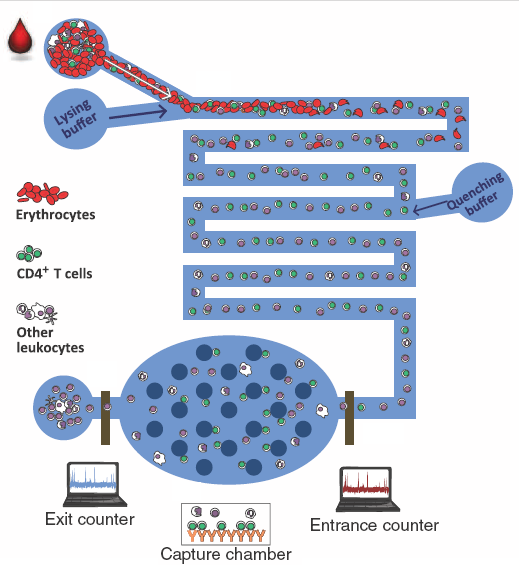
The blood cell count is one of the most ubiquitous tests performed in health care settings for disease diagnostics and its management. Irregular leukocyte and its differential count are observed in many diseases including leukemia, immunodeficiency, bone marrow failure, etc. CD4+ T cell count is used as a diagnostic test for HIV/AIDS and is also used for monitoring the effectiveness of antiretroviral therapy drugs on HIV infected patients. Similarly, the cancer patients undergoing chemo/radiation therapy suffer with very low total leukocyte count in particular absolute neutrophil count. The current standard to obtain cell counts requires a trained technician to operate a piece of large and expensive equipment i.e. flow cytometer. We have developed an immune-capture assay to electrically enumerate specific leukocytes in a microfluidic biochip with a drop of blood.
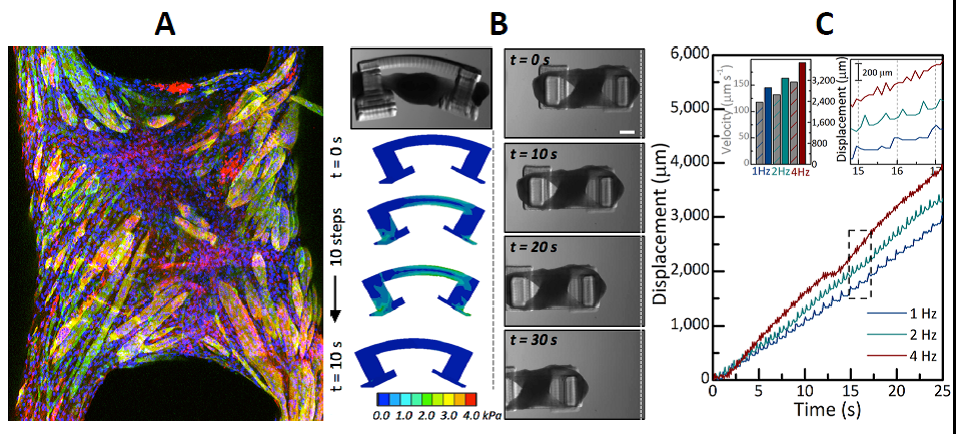
Ten microliters of whole blood are infused in biochip with the lysing buffer (composed of saponin and formic acid) to lyse all erythrocytes. To preserve the remaining leukocytes, the lysed solution is quenched to halt the lysing process and maintain the pH of the solution. The leukocytes are electrically counted as they pass through the counting channel bonded with microfabricated platinum electrodes. The specific leukocytes, e.g. CD4+T cells or neutrophils are captured in their respective capture chambers where their respective CD4 and CD66b antibody is adsorbed. The remaining cells are counted at the exit of the capture chamber by a second electrical counter. The difference in cell counts of two counters gives the CD4+ T cell count and absolute neutrophil count. The figure shows the comparison of CD4 T cell counts from HIV infected individuals using our biochip and control counts obtained from Carle Foundation Hospital.
A future goal of the project is to develop a biochip capable of performing a complete blood cell count. Our main focus is to perform an absolute count of red blood cell, platelets, white blood cells and its 3-part differential (Lymphocytes, Monocytes, and Neutrophils) from a drop of blood (finger-prick).
Characterization of Single-Cell Mass Response
 A complex relationship exists between the behavior of a cell and its physical properties. There is a strong interest in understanding the mechanisms through which the physical properties of a cell influence growth, differentiation, cell cycle progression, and apoptosis. Nanobiomechanics, which is the measuring of material properties at the smallest length scale, is an emerging area of research that has the ability to evolve the study of human disease. This field has helped elucidate mechanisms of disease progression and provided additional tools in the fight against the disease.
A complex relationship exists between the behavior of a cell and its physical properties. There is a strong interest in understanding the mechanisms through which the physical properties of a cell influence growth, differentiation, cell cycle progression, and apoptosis. Nanobiomechanics, which is the measuring of material properties at the smallest length scale, is an emerging area of research that has the ability to evolve the study of human disease. This field has helped elucidate mechanisms of disease progression and provided additional tools in the fight against the disease.
Much of the research on cell growth focuses on the temporal dynamics of growth rate. Variations in growth rate over the cell cycle may elucidate mechanisms underlying cell growth better than the magnitude of growth rate alone. There are two major models used to analyze the cell cycle: one based on an exponential increase and another based on a linear increase. The exponential growth rate for an individual cell is proportional to cell size mass, volume, or density during the cell cycle. The linear growth rate for an individual cell is constant meaning the cell increases size by the same amount regardless of its current size or state.
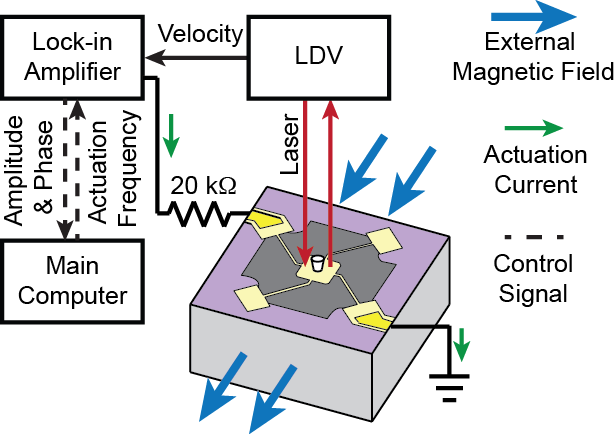
We developed an improved MEMS resonator sensor that can be used to directly measure the biophysical properties, mass, and growth rate of single adherent cells. These devices detect mass through a resonant shift and are able to be operated in a gaseous or aqueous environment. In order to overcome damping from the surrounding fluid, these devices must be driven magnetically to achieve a distinguishable resonance peak. However, our measurement technique offers a combination of complex elastic and viscoelastic dynamic properties of cells. Decoupling the relationship between the cell’s dynamics and the apparent mass reported by the sensor is of utmost importance. Understanding this relationship will further empower the measurement technique, enabling even more prudent investigations that will benefit efforts in cancer diagnosis and treatment, biological accurate design, cell-to-cell interfacing, and tissue engineering, among others.
Design and Fabrication of 3D Printed Biobots
 Over the past decade, a new scientific discipline has emerged, integrating mechanics with biology to create complex engineered living systems. The building blocks, different cell types in an instructive environment, can be assembled in various ways to promote the emergence (or natural evolution and interaction) of the cells in a system with well-defined functionality. These functions could include sensing, information processing, protein expression, and actuation.
Over the past decade, a new scientific discipline has emerged, integrating mechanics with biology to create complex engineered living systems. The building blocks, different cell types in an instructive environment, can be assembled in various ways to promote the emergence (or natural evolution and interaction) of the cells in a system with well-defined functionality. These functions could include sensing, information processing, protein expression, and actuation.
Scientists at the forefront of this research envision the extent of cellular and biological systems to include devices that are able to sense their surroundings and perform accordingly. Examples of cellular systems that can have real-world implications in medicine and the environment include:
- a biological “robot” or organism that has the ability to recognize a chemical gradient and produce a response, such as moving towards an oil spill for environmental remediation or neutralizing a dangerous toxin within the body;
- an engineered “organ” to sense drug and protein levels or glucose concentration in the bloodstream and send the information to a feedback loop or insulin pump;
- a micro-fluidic system acting as a human organ platform (heart, lung, or liver to eliminate harmful and costly drug development and testing; and
- plants with neurons that can send out a signal for water or nutrients after sensing a local deficiency.
All of these – and countless more – could potentially be achieved with the use of a few cell types, specifically neurons, endothelial cells, and muscle cells, which can be differentiated from stem cells.
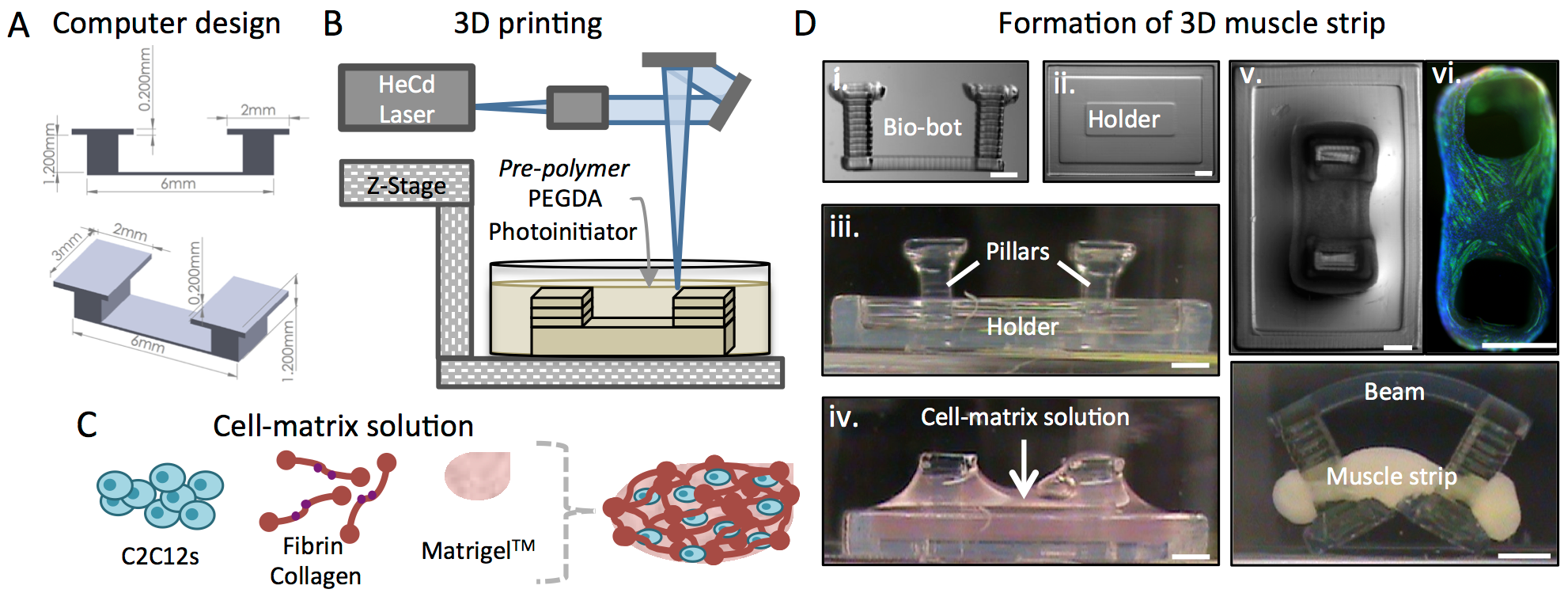
Cell-based soft robotic devices could have a transformative impact on our ability to design machines and systems that can dynamically sense and respond to a range of complex environmental signals. We demonstrate an interdisciplinary and novel integration of biomaterials, tissue engineering, and 3D printing to forward engineer controllable centimeter-scale biological machines capable of locomotion. Due in part to their elastic nature and the living components that can permit a dynamic response to environmental and applied stimuli, these biological machines can have diverse applications and represent a significant advancement towards high-level functional control over soft bio-robotic systems.
Combining biological components such as cells and tissues with soft robotics can enable the fabrication of biological machines with the ability to sense, process signals, and produce force. An intuitive demonstration of a biological machine is one that can produce motion in response to controllable external signaling. While cardiac cell-driven biological actuators have been demonstrated, the requirements of these machines to respond to stimuli and exhibit controlled movement merit the use of skeletal muscle, the primary generator of actuation in animals, as a contractile power source. We are interested in developing 3D printed hydrogel ‘bio-bots’ powered by the actuation of engineered mammalian skeletal muscle to result in net locomotion of the bio-bot.
The “biological parts” comprising many higher-level systems have been assembled to date as individual machine components with one or two cell types. Muscle cells have been used for biological actuators and neurons or neuronal networks assemble into sensory clusters or processors. The challenge, then, is to coordinate the cell types in a way that allows for new functionalities and applications in various fields. A complete cellular system needs many different components in order to be self-sustaining and autonomous: muscle cells that allow for movement, neuromuscular junctions to interface between muscle and neurons for control and processing, a vasculature supported by endothelial cells to provide nutrients and oxygen, and an exoskeleton for long-term viability and protection.
3D Microfabrication of Biological Machines
Projection Micro Stereolithography
Key Research Aims and Goals
Engineering in vitro model systems that recreate the structure and function of native tissue requires the ability to pattern cells and cell signals at physiologically relevant length scales.
This motivates the development of a 3D printing apparatus capable of patterning celss at high resolution, on the order of single cells (1-50 micron).
Research Highlights and Results
We have developed a custom-built projection micro-stereolithography apparatus capable of viably patterning cells encapsulated in hydrogel polymers at resolution < 5 micron.
The ability of this apparatus to fabricate multi-material biological structures with complex 3D designs is being used to target application in tissue engineering of neo-vasculature.
Biological Machines
Key Research Aims and Goals
Using enabling technology of 3D, we can forward engineer multi-functional integrated cellular systems that harness the innate dynamic abilities of cells to self-organize, self-heal, and respond to complex environmental cues.
Research Highlights and Results
We have developed a skeletal muscle-powered biological soft robotic devices or a bio-bot capable of controlled directional locomotion in response to external electrical signals.