Information about the Leckband Research Group.
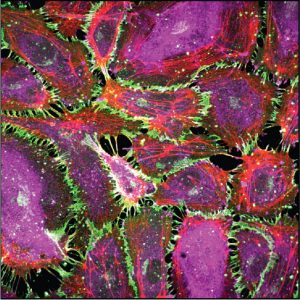 The Leckband Research Group focuses on biology at interfaces. It builds on our extensive expertise in nanoscale studies of biomaterial interfaces, immobilized proteins, and cell adhesion proteins. More than 85% of drugs target receptors on cell surfaces. Cells communicate with their environments through receptors on cell surfaces. Biosensor and biomaterials design exploit molecular recognition at solid/liquid interfaces. Yet interfacial environments differ radically from bulk solutions where most biomacromolecules are studied. This raises several questions: What’s special about interfaces? How can we study the interfacial properties that affect biological functions or device performance? How can we use our knowledge to improve human health? Our research builds on our extensive expertise in nanoscale studies of biomaterial interfaces, immobilized proteins, and cell adhesion proteins.
The Leckband Research Group focuses on biology at interfaces. It builds on our extensive expertise in nanoscale studies of biomaterial interfaces, immobilized proteins, and cell adhesion proteins. More than 85% of drugs target receptors on cell surfaces. Cells communicate with their environments through receptors on cell surfaces. Biosensor and biomaterials design exploit molecular recognition at solid/liquid interfaces. Yet interfacial environments differ radically from bulk solutions where most biomacromolecules are studied. This raises several questions: What’s special about interfaces? How can we study the interfacial properties that affect biological functions or device performance? How can we use our knowledge to improve human health? Our research builds on our extensive expertise in nanoscale studies of biomaterial interfaces, immobilized proteins, and cell adhesion proteins.
Our Principal Investigator is Deborah E. Leckband.
Check out the tabs below to learn about our research projects or here to visit our website.
Engineered Cell Environments
Lung on a Chip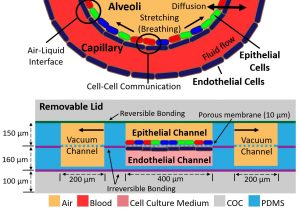
The increased use of nanoparticles in consumer products and therapeutic agents, and the prevalence of nanoparticles as environmental toxins increases concern about the adverse effects of nanoparticles on human health, especially the lung. Lung-on-a-chip microfluidic platforms possess the capability to mimic physiologically relevant lung tissue. The goal of my project is to use a sophisticated lung-on-a-chip platform to elucidate mechanisms of nanoparticle-induced inflammation and nanoparticle migration through lung tissue.
Neural Engineering
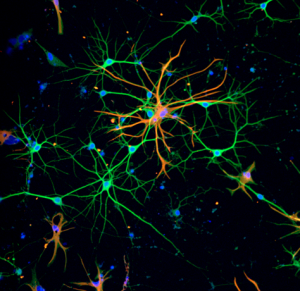 Neural (N-) cadherin is a crucial adhesion protein in the brain. N-cadherin regulates the functional organization of mature neural tissue and the formation of neural networks, by guiding axon extensions, neurite outgrowth, dendritic arborizations, and synaptogenesis. Studies are developing biomaterials that exploit N-cadherin to repair neural tissues in vivo. A broad goal is to engineer a brain in a dish to investigate mechanisms of neural development and neurodegenerative diseases.
Neural (N-) cadherin is a crucial adhesion protein in the brain. N-cadherin regulates the functional organization of mature neural tissue and the formation of neural networks, by guiding axon extensions, neurite outgrowth, dendritic arborizations, and synaptogenesis. Studies are developing biomaterials that exploit N-cadherin to repair neural tissues in vivo. A broad goal is to engineer a brain in a dish to investigate mechanisms of neural development and neurodegenerative diseases.
Biomaterials
Molecular Force Measurements
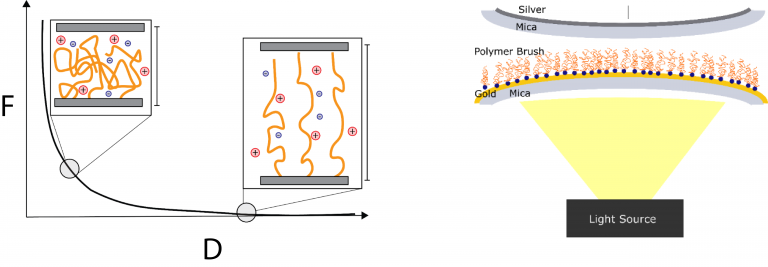
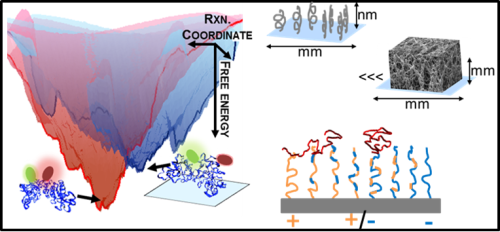
Nanoscale force measurements are used to quantify interfacial force fields that control biomaterial performance in medical applications. Studies focus on protein-compatible, water-soluble polymers used in drug delivery, tissue engineering, and biosensor coatings. Surface force apparatus (SFA) and colloidal probe atomic force microscopy (CP-AFM) measurements directly quantify force fields within 0.1-100 nm of material surfaces (as exemplified in figure 1). Current studies of Zwitterionic polymers (e.g. poly sulfobetaine) are identifying molecular design rules underlying the unique properties of these materials.
Biomaterial Effects on Protein Folding
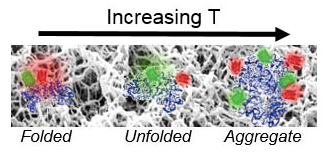
 The diversity of protein functions has inspired their use in hybrid bio-materials including biosensors, pharmaceuticals, tissue engineering, and drug targeting. Yet incorporating proteins in these abiotic environments can degrade their stability and function (see figure 2). Currently, few approaches can determine how material environments alter protein stability.
The diversity of protein functions has inspired their use in hybrid bio-materials including biosensors, pharmaceuticals, tissue engineering, and drug targeting. Yet incorporating proteins in these abiotic environments can degrade their stability and function (see figure 2). Currently, few approaches can determine how material environments alter protein stability.
We are developing Fast Relaxation Imaging (FReI) in collaboration with the Gruebele research group to define material design rules that preserve protein stability and function. FReI was developed to study protein folding in cells but has untapped potential to interrogate proteins in materials at sub-micron resolution. We are extending the FReI approach to image protein folding stability material. Studies of proteins in a “library” of materials are identifying novel, nanoscale design rules that preserve or shut down protein function.
We used FReI to determine how hydrogel–protein interactions and confinement in hydrogel nanopores alter protein folding stability.
Zwitterionic polymer-protein interactions
 Poly(zwitterions) are polymers that contain equal numbers of cationic and anionic groups on each monomer. They have remarkable anti-fouling properties and have potential biomedical applications in wound dressings, biosensors, or stealth nanoparticles for drug delivery. Poly(zwitterion) performance in these applications is ascribed to polymer solvation, which is postulated to prevent protein adsorption.
Poly(zwitterions) are polymers that contain equal numbers of cationic and anionic groups on each monomer. They have remarkable anti-fouling properties and have potential biomedical applications in wound dressings, biosensors, or stealth nanoparticles for drug delivery. Poly(zwitterion) performance in these applications is ascribed to polymer solvation, which is postulated to prevent protein adsorption.
This raises two very important questions:
- “Do poly(zwitterions) bind proteins?”
- “How do the polymers affect protein stability?”
We are studying poly(zwitterion) interactions with proteins both in solution and at surfaces. Fluorescence and biophysical measurements determine whether poly(zwitterions) bind proteins and the impact of protein-polymer interactions on protein stability and function. Our studies are identifying the physical-chemical bases for the success or failure of poly(zwitterionic) materials in diverse applications.
Cell Adhesion Biophysics
Intercellular Adhesion
Cell surface adhesion proteins are sites of force transmission in tissues. They maintain the organization of all multicellular organisms, regulate cell shape, maintain tissue barriers, and control immune cell surveillance. Cadherins are the biochemical Velcro that holds cells together in all tissues. We focus on how these essential intercellular adhesion proteins, cadherins interact with other proteins at the cell membrane and in the cytosol to regulate cell-cell adhesion, force transduction, and tissue physiology.
Receptor Binding Equilibria at Cell Surfaces
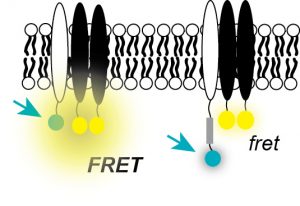
Fluorescence Analysis of Receptor Binding Equilibria
We discovered that cadherin-mediated force transduction at cell-cell adhesions activates growth factor receptor signaling and downstream pathways that regulate proliferation and differentiation. We are exploring the molecular basis of the interactions between cadherins and growth factor receptors, using a novel fluorescence technique FSI-FRET, developed by Kalina Hrisova at Johns Hopkins. These fluorescence measurements quantify the receptor binding stoichiometry and the binding affinities between membrane receptors at the surfaces of live cells. Related studies investigate how cadherin-mediated force transduction regulates cell proliferation and tissue morphogenesis, particularly in the contexts of vascular disease and breast cancer.
Kinetics and Mechanisms of Cell Adhesion
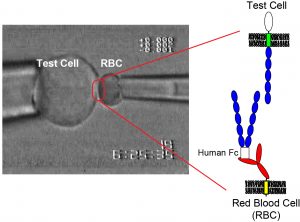
Our lab uses quantitative measurements of cell-cell binding kinetics to determine the molecular mechanisms of intercellular adhesion and molecular organization at intercellular junctions. Single-molecule force measurements quantified the strengths of single protein-ligand bonds. These measurements in conjunction with super-resolution imaging, protein engineering, and kinetic modeling reveal how adhesion proteins control the assembly and function of the intercellular adhesions, which determine the permeability of barrier tissues such as skin and the vascular endothelium.
Mechanotransduction from atoms to tissues
Mechanotransduction
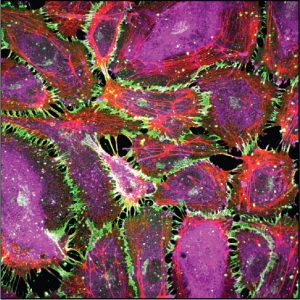
We are determining how cells convert mechanical cues into signals that regulate cell proliferation, differentiation and tissue functions. Studies investigate mechanotransduction across length scales, from atomistic simulations to 3D organoids. We develop engineering and biophysical tools to determine how mechanical stimuli in vivo such as cyclic stretch in lungs regulate tissue functions. Research focuses primarily on intercellular adhesion proteins cadherins, which are critical mechanical and signaling hubs in all tissues. We further identify the clinical significance of force transduction in vascular disease and breast cancer and exploiting these findings to regulate tissue functions. We collaborate with clinicians and cell biologists at UCSF, Univ of Washington, Northwestern, Univ of Toronto, UI Chicago, and UIUC.
Molecular force transduction
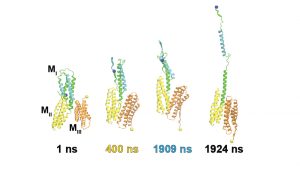
MD simulations, dynamic fluorescence imaging using protein biosensors, and biochemical approaches are identifying molecular mechanisms of force transduction in living cells. Simulations reveal how force alters protein conformations (and function) to activate intracellular signals (force transduction). We test model predictions using live-cell fluorescence imaging and cell engineering. Dynamic fluorescence imaging, quantitative immunofluorescence, and cell engineering reveal force activated molecular cascades in live cells. Further studies assess the impact of these molecular-scale events on cell and tissue functions.