Information about the Bell Lab.

Research in the Bell lab is focused on understanding why individual animals behave differently from each other. Even an individual fish, for example, behaves differently from other fish, through time and across situations. We study the proximate and ultimate causes of individual variation in the three-spined stickleback. Work in the Bell lab is focused on understanding behavioral variation both from a proximate and ultimate perspective. To that end, we study the neuroendocrine and molecular mechanisms that contribute to behavior, and the selective pressures that maintain variation within natural populations.
The Bell lab is involved in a collaborative project in our theme at the Carl Woese Institute for Genomic Biology to test the hypothesis that there is a ‘genetic toolkit’ for social behavior. We are comparing neurogenomic responses to social opportunities and challenges among honeybees, sticklebacks, and mice (PIs: Gene Robinson and Lisa Stubbs).
Our Principal Investigator is Alison Marie Bell.
Check out at our website here.
Why Study Three-Spined Stickleback?
We explore two questions in three-spined sticklebacks: Why do individuals vary? Why do individuals behave consistently? Three-spined sticklebacks are a small fish that have proven to be a powerful model system for identifying the genetic mechanisms that underlie adaptive morphological evolution. However, sticklebacks are also famous for their rich behavioral repertoire, which has attracted the attention of biologists since the days of the early ethologists, including Nobel Prize-winner Niko Tinbergen. For example, male sticklebacks aggressively defend nesting territories and are the sole providers of parental care that is necessary for offspring survival. Some individual sticklebacks are more willing to take risks than others, and the way an individual stickleback behaves toward competitors, predators, mates, and offspring directly influences fitness and has a heritable component. Many of the axes of natural behavioral variation studied in sticklebacks have parallels with human behavioral variation, e.g. risk-taking behavior, sensation seeking, extraversion, and aggressiveness. Moreover, the system is tractable: sticklebacks can be readily drawn from the wild and measured in reliable behavioral assays, large sample sizes are feasible and they can be crossed and reared in the lab. In other words, sticklebacks have many of the advantages of traditional model organisms with the added advantage that we can study them in a wild state.
Present work in the lab focuses on behavioral variation, behavioral plasticity, transgenerational plasticity, and individual differences. Our work combines field and lab behavioral experiments, neuroendocrine analyses and often capitalizes on the natural diversity among different populations to understand the evolutionary context in which behavioral variation and plasticity play out. Many of the current projects in the lab use neurogenomic techniques, including measuring brain gene expression with RNA-seq and relating changes in gene expression to changes in chromatin accessibility with ChIP-seq in order to understand how the genome dynamically responds to the environment (Bell and Robinson 2011). Our lab performed the first brain gene expression studies on this species when we asked how the expression of the genome changes in response to predation risk (Sanogo et al. 2011). Since then, we’ve studied how the genome responds to a territorial challenge (Sanogo et al. 2012, Bukhari et al. 2017), a courtship opportunity (Sanogo and Bell 2016), both mothers’ (Mommer and Bell 2014) and fathers’ (Stein and Bell 2014) experience with predation risk, and how brain gene expression profiles differ between behavioral types (Bell et al. 2016). To help understand the functional relationships between gene expression and behavior, we are developing viral-mediated gene transfer methods in the brain as part of a collaborative NSF EDGE grant to develop functional tools for sticklebacks. Check out the tabs below to learn more
Transgenerational Plasticity
 Over the past several years the lab has grown increasingly interested in the possibility that the environment experienced by parents can influence the phenotypic development of their offspring (transgenerational plasticity). We have shown that female sticklebacks are an important source of variation in their offspring, not just via their genes, but via the environment, they provide for their offspring. In particular, we found that mothers exposed to threatening stimuli such as a predator produce offspring with altered behavior (Giesing et al 2011), stress response (Giesing et al 2011) and learning (Roche et al 2012). When testing the hypothesis that predator-mediated maternal effects are mediated through the glucocorticoid stress response system, Ryan Paitz discovered that embryos are capable of buffering themselves from maternal steroids by actively transporting them out of the egg (Paitz et al 2016). Katie McGhee – a former postdoc in the lab – also showed that fathers change their paternal behavior when mated to predator-exposed females (McGhee et al 2015), illustrating the dynamic interplay between mothers and fathers.
Over the past several years the lab has grown increasingly interested in the possibility that the environment experienced by parents can influence the phenotypic development of their offspring (transgenerational plasticity). We have shown that female sticklebacks are an important source of variation in their offspring, not just via their genes, but via the environment, they provide for their offspring. In particular, we found that mothers exposed to threatening stimuli such as a predator produce offspring with altered behavior (Giesing et al 2011), stress response (Giesing et al 2011) and learning (Roche et al 2012). When testing the hypothesis that predator-mediated maternal effects are mediated through the glucocorticoid stress response system, Ryan Paitz discovered that embryos are capable of buffering themselves from maternal steroids by actively transporting them out of the egg (Paitz et al 2016). Katie McGhee – a former postdoc in the lab – also showed that fathers change their paternal behavior when mated to predator-exposed females (McGhee et al 2015), illustrating the dynamic interplay between mothers and fathers.
 Sticklebacks are particularly interesting subjects for studying transgenerational plasticity mediated by parental behavior because fathers provide care for their developing offspring, and we’ve been interested in the possibility that the early environmental provided by fathers is important for offspring development. When she was a Ph.D. student in the lab, Laura Stein found that fathers change the way they parent in response to predation risk (Stein and Bell 2014) and that offspring of predator exposed fathers are programmed to respond to danger (Stein and Bell 2014). Moreover, the effects of paternal experience with danger resemble the effects of personal, early life experience with danger (Stein et al 2018). Jason Keagy is actively investigating the epigenetic mechanisms by which parents’ experiences can influence the next generation (McGhee and Bell 2014). Jenn Hellmann is currently studying sperm-mediated paternal effects, and their consequences for multiple generations, and we recently published a paper articulating a new framework for understanding the mechanisms and multigenerational consequences of transgenerational plasticity (Bell and Hellmann 2019).
Sticklebacks are particularly interesting subjects for studying transgenerational plasticity mediated by parental behavior because fathers provide care for their developing offspring, and we’ve been interested in the possibility that the early environmental provided by fathers is important for offspring development. When she was a Ph.D. student in the lab, Laura Stein found that fathers change the way they parent in response to predation risk (Stein and Bell 2014) and that offspring of predator exposed fathers are programmed to respond to danger (Stein and Bell 2014). Moreover, the effects of paternal experience with danger resemble the effects of personal, early life experience with danger (Stein et al 2018). Jason Keagy is actively investigating the epigenetic mechanisms by which parents’ experiences can influence the next generation (McGhee and Bell 2014). Jenn Hellmann is currently studying sperm-mediated paternal effects, and their consequences for multiple generations, and we recently published a paper articulating a new framework for understanding the mechanisms and multigenerational consequences of transgenerational plasticity (Bell and Hellmann 2019).
Paternal Care
 Parental care is fascinating for countless reasons – it’s critical for fitness, there is tremendous variation in the specific forms and manifestations of parental care across organisms, and it’s a highly dynamic interaction between relatives. One of the things that make sticklebacks – like many other fishes – interesting is that they exhibit paternal care, where the father is solely responsible for the care of the offspring. We are very interested in the evolution, inheritance, neurogenomics, and plasticity of paternal care in sticklebacks.
Parental care is fascinating for countless reasons – it’s critical for fitness, there is tremendous variation in the specific forms and manifestations of parental care across organisms, and it’s a highly dynamic interaction between relatives. One of the things that make sticklebacks – like many other fishes – interesting is that they exhibit paternal care, where the father is solely responsible for the care of the offspring. We are very interested in the evolution, inheritance, neurogenomics, and plasticity of paternal care in sticklebacks.
 We’ve found that some male sticklebacks provide more care than others (Stein 2012, Stein and Bell 2015), the quantity of care received influences offspring traits (McGhee and Bell 2014), and that paternal care might have contributed to the stickleback radiation (Stein and Bell 2019). In a recent study, we found that individual differences in a key parental behavior, fanning, is highly heritable among individual male sticklebacks from the same population (Bell et al 2018). These results suggest that variation in paternal care is tractable for further genetic dissection, e.g. via QTL or GWAS. Colby Behrens (Ph.D. student in Evolution, Ecology, and Behavior) is capitalizing on natural variation among stickleback populations in paternal care (including the so-called “white sticklebacks” in Nova Scotia that do not provide care) in order to understand more about its genetic basis.
We’ve found that some male sticklebacks provide more care than others (Stein 2012, Stein and Bell 2015), the quantity of care received influences offspring traits (McGhee and Bell 2014), and that paternal care might have contributed to the stickleback radiation (Stein and Bell 2019). In a recent study, we found that individual differences in a key parental behavior, fanning, is highly heritable among individual male sticklebacks from the same population (Bell et al 2018). These results suggest that variation in paternal care is tractable for further genetic dissection, e.g. via QTL or GWAS. Colby Behrens (Ph.D. student in Evolution, Ecology, and Behavior) is capitalizing on natural variation among stickleback populations in paternal care (including the so-called “white sticklebacks” in Nova Scotia that do not provide care) in order to understand more about its genetic basis.
However, paternal care is not only highly heritable among individuals but it is also highly plastic within individuals: paternal care changes not only over the course of the breeding cycle but in immediate response to threatening stimuli (Stein and Bell 2014). Moreover, Abbas Bukhari et al showed that males undergo dramatic changes in brain gene expression as they become parents, and there are fascinating neurogenomic commonalities between paternal care in a fish and maternal care in a mouse (Bukhari et al in press)!
Behavioral Genomics
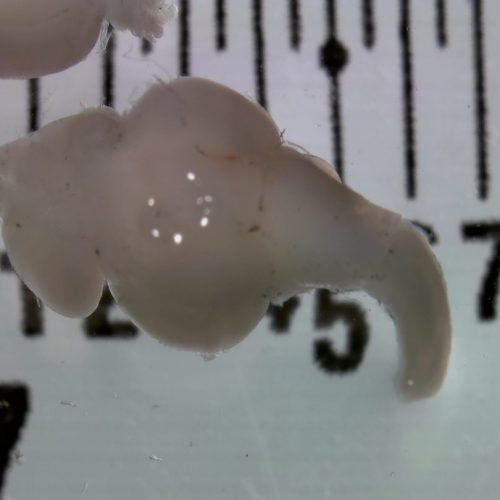 Behavioral traits offer exciting new opportunities for integrating quantitative genetics with results from studies of molecular mechanisms and genomics (Bell and Dochtermann 2015). The rationale is that in order to fully understand behavioral traits, they require consideration of both variation and plasticity, and the fixed and dynamic sides of the genome. The growing field of animal personality research prompts this integration because it has brought questions about how and why variation and plasticity coexist to the forefront of thinking in evolutionary and behavioral ecology. We would argue, moreover, that these features of behavioral traits (variation among individuals and plasticity within individuals) are shared with other fitness-related traits of interest to biologists, including health-related and physiological traits, therefore our findings have broad relevance (Bengston et al 2018).
Behavioral traits offer exciting new opportunities for integrating quantitative genetics with results from studies of molecular mechanisms and genomics (Bell and Dochtermann 2015). The rationale is that in order to fully understand behavioral traits, they require consideration of both variation and plasticity, and the fixed and dynamic sides of the genome. The growing field of animal personality research prompts this integration because it has brought questions about how and why variation and plasticity coexist to the forefront of thinking in evolutionary and behavioral ecology. We would argue, moreover, that these features of behavioral traits (variation among individuals and plasticity within individuals) are shared with other fitness-related traits of interest to biologists, including health-related and physiological traits, therefore our findings have broad relevance (Bengston et al 2018).
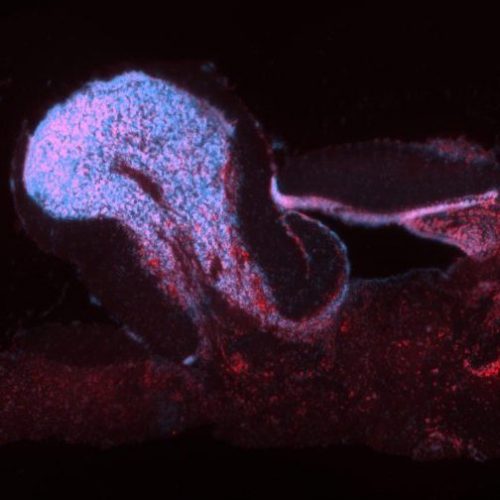 When our lab was established, the stickleback genome had just been sequenced and we saw opportunities to use emerging genomic technologies to uncover the proximate causes of individual differences in behavior. A genome-wide approach was appealing because the behaviors of interest are influenced by many interacting genes of small effect and candidate genes for the behaviors explained only a small fraction of the total variation. Moreover, measuring gene expression (rather than quantifying static DNA sequence variation) was attractive because while there is a heritable component to behavioral traits, they are also plastic, and change with the environment. Instead of focusing on the static side of the genome (DNA sequence), initial studies in the lab probed behavioral variation at the molecular level by measuring brain genome-wide expression in order to learn how the genome dynamically responds to the environment. We found that the stickleback genome is highly responsive to many important aspects of the environment such as predation risk (Sanogo et al 2011), a territorial intrusion (Bukhari et al 2017) and a courtship opportunity (Sanogo and Bell 2016). An important element of this line of work includes tool development for this system. Noelle James published a method for in situ hybridization in stickleback brain tissue (James et al 2016) and is currently developing viral-mediated gene transfer methods in this system as part of an NSF EDGE grant.
When our lab was established, the stickleback genome had just been sequenced and we saw opportunities to use emerging genomic technologies to uncover the proximate causes of individual differences in behavior. A genome-wide approach was appealing because the behaviors of interest are influenced by many interacting genes of small effect and candidate genes for the behaviors explained only a small fraction of the total variation. Moreover, measuring gene expression (rather than quantifying static DNA sequence variation) was attractive because while there is a heritable component to behavioral traits, they are also plastic, and change with the environment. Instead of focusing on the static side of the genome (DNA sequence), initial studies in the lab probed behavioral variation at the molecular level by measuring brain genome-wide expression in order to learn how the genome dynamically responds to the environment. We found that the stickleback genome is highly responsive to many important aspects of the environment such as predation risk (Sanogo et al 2011), a territorial intrusion (Bukhari et al 2017) and a courtship opportunity (Sanogo and Bell 2016). An important element of this line of work includes tool development for this system. Noelle James published a method for in situ hybridization in stickleback brain tissue (James et al 2016) and is currently developing viral-mediated gene transfer methods in this system as part of an NSF EDGE grant.
Much of the behavioral genomics work in the lab is carried out in collaboration with our theme at the Carl Woese Institute for Genomic Biology and is currently being spearheaded by Abbas Bukhari. We are comparing neurogenomic responses to social opportunities and challenges among honeybees, sticklebacks, and mice (Rittschof et al 2014, Saul et al 2019).
Individual Differences
 A theme running throughout all of the work in the lab has to do with individual variation. Almost 20 years ago, I had the privilege of working with Andy Sih and colleagues to develop a framework for understanding the evolutionary and ecological causes and significance of behavioral variation and plasticity (Sih et al 2004a,b), which now goes by the name of behavioral syndromes and animal personality. As a Ph.D. student working with Judy Stamps, I revisited Felicity Huntingford’s classic work on individual differences in boldness and aggression in sticklebacks (Huntingford 1976), and showed that the boldness-aggressiveness behavioral syndrome is not invariant, but it can vary over both developmental (Bell and Stamps 2004) and evolutionary timescales (Bell 2005), consistent with the hypothesis that behavioral variation and covariation is adaptive (Bell and Sih 2007). The Bell lab has continued to make important contributions to this area, including a meta-analysis of repeatability estimates (Bell et al 2009). Miles Bensky is currently studying individual differences in behavioral flexibility and persistence in sticklebacks.
A theme running throughout all of the work in the lab has to do with individual variation. Almost 20 years ago, I had the privilege of working with Andy Sih and colleagues to develop a framework for understanding the evolutionary and ecological causes and significance of behavioral variation and plasticity (Sih et al 2004a,b), which now goes by the name of behavioral syndromes and animal personality. As a Ph.D. student working with Judy Stamps, I revisited Felicity Huntingford’s classic work on individual differences in boldness and aggression in sticklebacks (Huntingford 1976), and showed that the boldness-aggressiveness behavioral syndrome is not invariant, but it can vary over both developmental (Bell and Stamps 2004) and evolutionary timescales (Bell 2005), consistent with the hypothesis that behavioral variation and covariation is adaptive (Bell and Sih 2007). The Bell lab has continued to make important contributions to this area, including a meta-analysis of repeatability estimates (Bell et al 2009). Miles Bensky is currently studying individual differences in behavioral flexibility and persistence in sticklebacks.